Abstract
Context. Mutations affecting cytokine signal transduction genes ('signaling' genes) are frequent in CBF AML and increase the risk of relapse. Clonal heterogeneity heralds poor prognosis in many tumors. Clonal interference is a specific type of clonal heterogeneity defined by the coexistence of independent clones affecting a common pathway, and has been linked to resistance to selection pressures in bacteria and viruses (Hughes, Nat Rev Genet 2015). Single-cell data has shown that in CBF AML with several signaling mutations, these mutations occur in independent clones (Hirsch, Nat Comm 2015). We investigated the prognostic role of clonal interference of signaling mutations in intensively-treated CBF AML.
Methods. Diagnostic bone marrow (BM) samples from CBF AML cases from 2 multicenter adult and pediatric trials underwent targeted high-throughput sequencing (HTS) in Lille (n=215) as reported (Duployez Blood 2016). Diagnostic samples from adult CBF AML cases referred to Munich Leukemia Laboratory were analyzed with an independent HTS platform, including previously published RUNX1 - RUNX1T1 cases (CBFA, Krauth, Leukemia 2014) and additional CBFB - MYH11 (CBFB) cases (n=230 in total). All analyses were stratified on the HTS platform. We defined the number of 'signaling clones' as the total number of variants found in signaling genes (KIT, FLT3, NRAS, KRAS, JAK2 and CBL) by HTS in each patient.
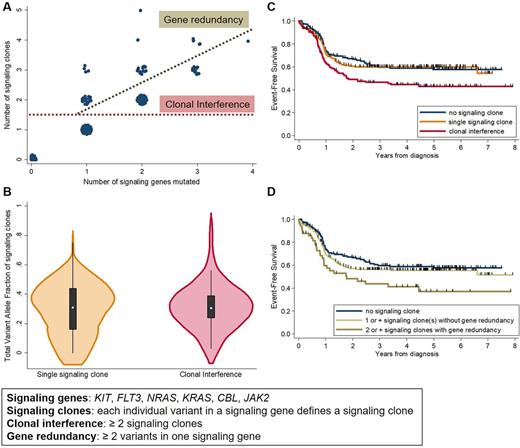
Results. The study cohort included 445 patients (pts, M/F 251/194, median age 42y [IQR 24 - 54]) with CBFA (n=230) and CBFB (n=215) AML. Median WBC was 14.6 x109/L. Sequencing data for all 445 pts was available for 6 signaling genes, with the following mutations frequencies: KIT 31%, NRAS 29%, FLT3 16% (TKD 10%, ITD 5%, both 1%), KRAS 13%, CBL (2%) and JAK2 (1%). FLT3-TKD, NRAS and KRAS mutations were significantly more frequent in CBFA AML, and FLT3-ITD in CBFB AML. Recurrent co-mutations in genes covered in >90% of cases included TET2 (6%) and ASXL1 (5%). At least one mutation in KIT, NRAS, KRAS, FLT3, CBL or JAK2 was found in 293 (66%) pts, of whom 126 harbored clonal interference, defined as ≥2 signaling clones, because of mutations in different genes (n=76), different variants in one gene (n=27), or both (n=23, Figure A). Gene redundancy, defined as the presence of ≥2 variants in one gene, particularly affected NRAS (n=27/50 cases) and KIT (n=12/50).
Among cases with at least one signaling clone, clonal interference was more frequent in CBFB AML (48% vs 36% in CBFA, p=0.025), but was not associated with age, gender, leukocytosis, or to mutations in ASXL1 or TET2 . Importantly, the total allelic burden of signaling clones was comparable in cases with and without clonal interference (p=0.2, Figure B).
All pts were treated with different 7+3 based intensive chemotherapy regimens as previously published (Duployez Blood 2016, Krauth Leukemia 2014). Median follow-up was 4.7 years, 47 (11%) pts received an allogeneic transplant in first CR. Censoring at transplant did not change our results. Three-year event-free survival (EFS) was 59.8% (95% CI, 51.2 - 67.3) in pts with no signaling mutation, 60.0% (52.0 - 67.2) in pts with a single signaling clone, and 46.4% (37.2 - 55.2) in pts with clonal interference (p=0.001, Figure C). This difference in EFS was seen in CBFB and to a lesser extent in CBFA AML. Clonal interference increased the cumulative incidence of failure (hazard ratio [HR]=1.54 compared to pts with a single signaling clone; p=0.03) translating into a trend towards poorer overall survival (HR=1.55; p=0.07). Martingale residuals indicated that each additional signaling clone worsened EFS (HR=1.30; p<0.0001). Presence of gene redundancy (≥2 variants in one gene) predicted worse EFS (p=0.04; Figure D) independently of the number of mutated signaling genes (p=0.10), further strengthening the adverse prognostic value of intratumoral heterogeneity in CBF AML. In a multivariate Cox model, clonal interference predicted a worse EFS (HR=2.32, p=0.002) independently of older age (p<0.0001), higher WBC (p<0.0001), CBF subtype (CBFA vs CBFB, p=0.004), and total allelic burden of signaling clones (p=0.13).
Conclusion. Our results in a large cohort of CBF AML patients demonstrate that presence of two or more genetic lesions in recurrently mutated cytokine signaling genes predicts poor prognosis in CBF AML independently of clinical features. Our analyses suggest that clonal interference contributes to chemoresistance in AML.
Itzykson: Janssen: Research Funding; Novartis: Research Funding. Fasan: MLL Munich Leukemia Laboratory: Employment. Meggendorfer: MLL Munich Leukemia Laboratory: Employment. Jourdan: NOVARTIS: Consultancy, Honoraria. Haferlach: MLL Munich Leukemia Laboratory: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal